Abstract – US National Library of Medicine & National Institutes of Health
By: Elizabeth A. Grice and Julia A. Segre
The skin is the human body’s largest organ, colonized by a diverse milieu of microorganisms, most of which are harmless or even beneficial to their host. Colonization is driven by the ecology of the skin surface, which is highly variable depending on topographical location, endogenous host factors and exogenous environmental factors. The cutaneous innate and adaptive immune responses can modulate the skin microbiota, but the microbiota also functions in educating the immune system. The development of molecular methods to identify microorganisms has led to an emerging view of the resident skin bacteria as highly diverse and variable. An enhanced understanding of the skin microbiome is necessary to gain insight into microbial involvement in human skin disorders and to enable novel promicrobial and antimicrobial therapeutic approaches for their treatment.
The skin is an ecosystem composed of 1.8 m2 of diverse habitats with an abundance of folds, invaginations and specialized niches that support a wide range of microorganisms. The primary role of the skin is to serve as a physical barrier, protecting our bodies from potential assault by foreign organisms or toxic substances. The skin is also an interface with the outside environment and, as such, is colonized by a diverse collection of microorganisms — including bacteria, fungi and viruses — as well as mites1-7 (FIG. 1). As we describe, many of these microorganisms are harmless and in some cases provide vital functions that the human genome has not evolved. Symbiotic microorganisms occupy a wide range of skin niches and protect against invasion by more pathogenic or harmful organisms. These microorganisms may also have a role in educating the billions of T cells that are found in the skin, priming them to respond to similarly marked pathogenic cousins.
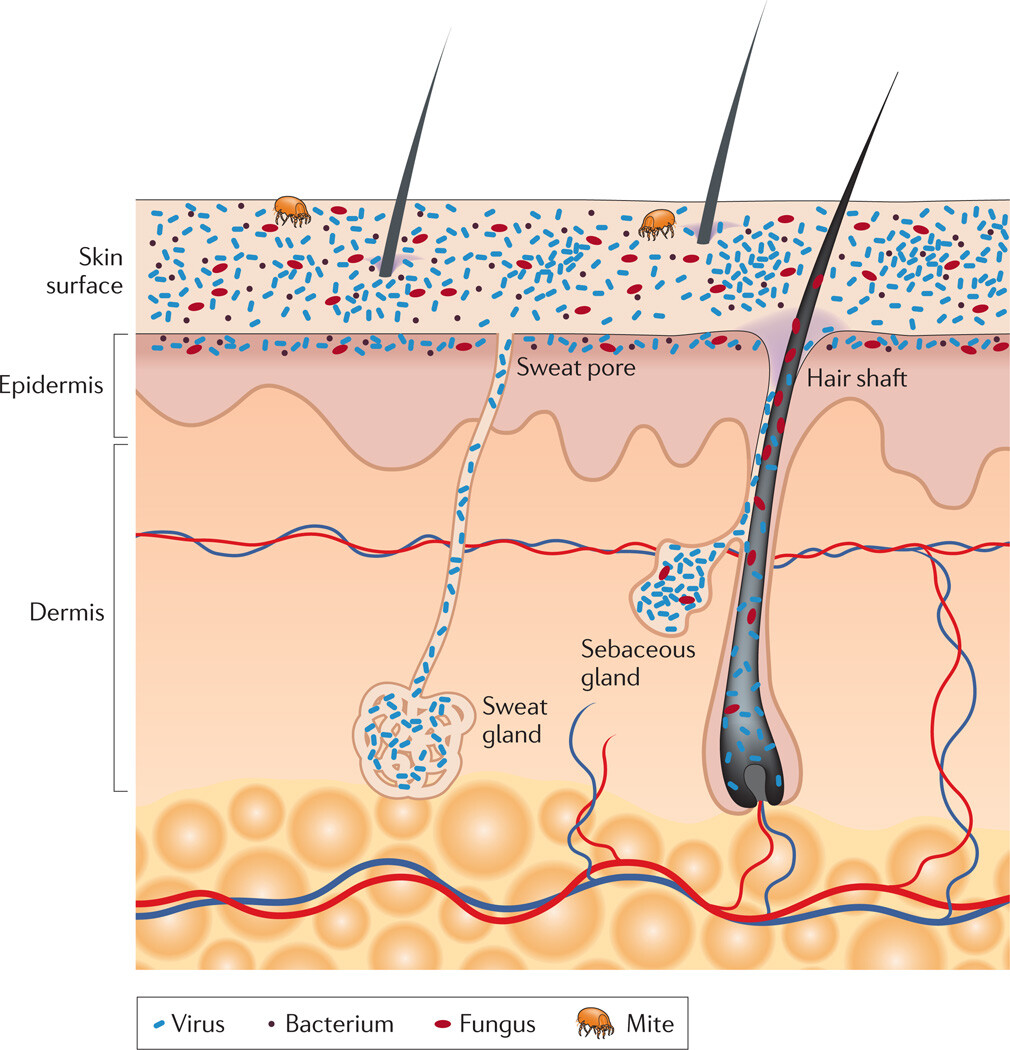
Figure 1
Schematic of skin histology viewed in cross-section with microorganisms and skin appendages
Microorganisms (viruses, bacteria and fungi) and mites cover the surface of the skin and reside deep in the hair and glands. On the skin surface, rod and round bacteria — such as Proteobacteria and Staphylococcus spp., respectively — form communities that are deeply intertwined among themselves and other microorganisms. Commensal fungi such as Malassezia spp. grow both as branching filamentous hypha and as individual cells. Virus particles live both freely and in bacterial cells. Skin mites, such as Demodex folliculorum and Demodex brevis, are some of the smallest arthropods and live in or near hair follicles. Skin appendages include hair follicles, sebaceous glands and sweat glands.
The perception of the skin as an ecosystem — composed of living biological and physical components occupying diverse habitats — can advance our understanding of the delicate balance between host and microorganism. Disruptions in the balance on either side of the equation can result in skin disorders or infections. Perturbations affecting the host–microorganism relationship can be endogenous (for example, genetic variation that selects for a specific microbial community) or exogenous (for example, hand washing). To further our understanding of health, disease and infection of the skin, microbiologists, immunologists and dermatologists have partnered with genomic scientists to develop a more complete characterization of the skin microbiota and how it interacts with the host (FIG. 2).
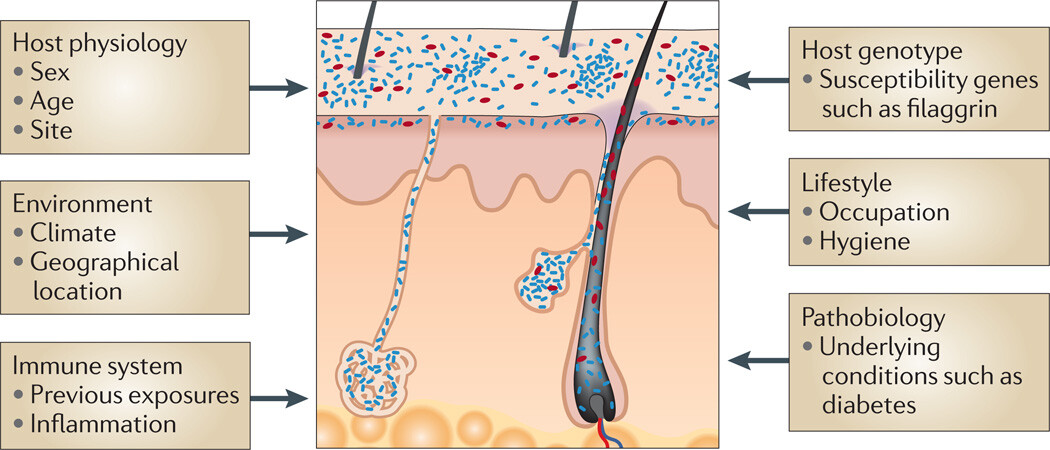
Figure 2
Factors contributing to variation in the skin microbiome
Exogenous and endogenous factors discussed in this Review that contribute to variation between individuals and over the lifetime of an individual.
The habitat of the skin defined
The physical and chemical features of the skin select for unique sets of microorganisms that are adapted to the niche they inhabit. In general, the skin is cool, acidic and desiccated, but distinct habitats are determined by skin thickness, folds and the density of hair follicles and glands8. Structurally, the epidermis is a formidable physical barrier, resisting penetration by microorganisms and potential toxins while retaining moisture and nutrients inside the body9-11. The top layer of the epidermis, the stratum corneum (FIG. 1), is composed of terminally differentiated, enucleated keratinocytes, that are known as squames. Squames consist of keratin fibrils and crosslinked, cornified envelopes embedded in lipid bilayers, forming the ‘bricks and mortar’ of the epidermis11. The skin is a continuously self-renewing organ, and squames are constantly shed from the skin surface as the final stage of terminal differentiation, having begun their migration from the basal layer ~4 weeks earlier12.
Invaginations and appendages
Cutaneous invaginations and appendages, including sweat glands (eccrine and apocrine), sebaceous glands and hair follicles, are likely to be associated with their own unique microbiota13 (FIG. 1). Eccrine glands, which are more abundant than apocrine glands, are found on virtually all skin surfaces and continuously bathe the skin surface with their secretion, which is composed mainly of water and salt. The primary role of eccrine sweat is thermoregulation through the release of latent heat from the evaporation of water. Additional functions of eccrine glands include excretion of water and electrolytes, and acidification of the skin, which prevents the colonization and growth of microorganisms. Apocrine glands, which are located in the axillary vault (armpit), nipple and genitoanal regions, respond to adrenaline by producing milky, viscous, odourless secretions. Apocrine secretions have long been postulated to contain pheromones, which are molecules that trigger certain behaviours (for example, sexual or alarm) in the receiving individual14. The stereotypical odour associated with sweat derives from bacterial processing and utilization of apocrine gland secretions15-18.
Sebaceous glands are connected to the hair follicle, forming the pilosebaceous unit, and secrete the lipid-rich substance sebum. Sebum is a hydrophobic coating that protects and lubricates the skin and hair and provides an antibacterial shield. Sebaceous glands are relatively anoxic and support the growth of facultative anaerobes such as Propionibacterium acnes, a common skin commensal bacterium3,13. Full genome sequencing of P. acnes has revealed multiple genes encoding lipases that degrade skin lipids of sebum19. P. acnes hydrolyses the triglycerides present in sebum, releasing free fatty acids onto the skin20,21. The bacterium can then adhere to these free fatty acids, and this perhaps aids in the colonization of the sebaceous gland22. These free fatty acids also contribute to the acidic pH (~5) of the skin surface4,10. Many common pathogens, such as Staphylococcus aureus and Streptococcus pyogenes, are inhibited by an acidic pH, thus the growth of coagulase- negative staphylococci and corynebacteria is favoured10,23–25. However, skin occlusion results in an elevated pH, which favours the growth of S. aureus and S. pyogenes24. Because humans produce much greater quantities of triglyceride-containing sebum than other mammals, P. acnes is present in greater abundance on human skin than on the skin of other mammals26.
Topography
The skin surface varies topographically owing to regional differences in skin anatomy and, according to culture-based studies, these regions are known to support distinct sets of microorganisms. Some regions of the skin are partially occluded, such as the groin, axillary vault and toe web. These regions are higher in temperature and humidity, which encourages the growth of microorganisms that thrive in moist conditions (for example, Gram-negative bacilli, coryneforms and S. aureus)4. The density of sebaceous glands is another factor that influences the skin microbiota, depending on the region. Areas with a high density of sebaceous glands, such as the face, chest and back, encourage the growth of lipophilic microorganisms (for example, Propionibacterium spp. and Malassezia spp.)4. Compared with other skin sites, arm and leg skin is relatively desiccated and experiences large fluctuations in surface temperature. Using culture-based methods, these areas were found to harbour quantitatively fewer organisms than moist areas of the skin surface3.
Host factors
Factors specific to the host, such as age, location and sex, contribute to the variability seen in the microbial flora of the skin (FIG. 2). Age has a great effect on the microenvironment of the skin and, thus, on the colonizing microbiota27,28. In utero, fetal skin is sterile, but colonization occurs immediately after birth, either during vaginal delivery or in the minutes following birth by caesarian section29,30. One area for future research is to explore how the microbial communities of the skin and other sites are established and stabilized during the first years of life, as a newborn baby explores its environment and matures its immune system31. During puberty, changes in sebum production parallel the levels of lipophilic bacteria on the skin, as determined by culture-based approaches28. Physiological and anatomical differences between male and female cutaneous environments such as sweat, sebum and hormone production, partially account for the microbial differences seen between the genders32–34.
Environmental factors
Environmental factors specific to the individual, such as occupation, clothing choice and antibiotic usage, may modulate colonization by the skin microbiota (FIG. 2). The effect of antibiotic treatment on the gut microbiota has been examined using molecular methods35–37 but, to our knowledge, a similar assessment of skin microbiota in healthy individuals does not exist. Cosmetics, soaps, hygienic products and moisturizers are also potential factors contributing to the variation of skin microbiota. These products alter the conditions of the skin barrier but their effects on skin microbiota remain unclear. Quantitative culture demonstrated that high-temperature and high-humidity are associated with increased quantities of bacteria on the back, axillary vaults and feet as compared with high-temperature low-humidity conditions38. In the same study, high humidity and low temperature conditions were associated with a higher frequency of Gram-negative bacteria on the back and feet. Ultraviolet (UV) light is a welld-ocumented bactericidal treatment39, and one can imagine geographical variability in skin microbiota correlating with the longitudinal and/or latitudinal variation in UV exposure.
Molecular analysis of skin microbiota
Genomic approaches to characterize skin bacteria have revealed a much greater diversity of organisms than that revealed by culture-based methods33,40–43 (BOX 1). As defined by 16S ribosomal RNA metagenomic sequencing, most skin bacteria fall into four different phyla: Actinobacteria, Firmicutes, Bacteroidetes and Proteobacteria. These four dominant phyla also constitute the microbiota that is found on the inner mucosal surfaces (the gastrointestinal tract and oral cavity44–49). However, the proportions differ vastly: whereas Actinobacteria members are more abundant on skin, Firmicutes and Bacteroidetes members are more abundant in the gastrointestinal tract. A common feature of gut and skin microbial communities seems to be low diversity at the phylum level, but high diversity at the species level.
Box 1 | A historical view of the skin microbiota
Microorganisms colonizing the skin have long been of interest to dermatologists and microbiologists; our knowledge of these microorganisms has, until recently, been gleaned through culture-based studies. Historically, Staphylococcus epidermidis and other coagulase-negative staphylococci have been regarded as the primary bacterial colonizers of the skin. Other microorganisms that are generally regarded as skin colonizers include coryneforms of the phylum Actinobacteria (the genera Corynebacterium, Propionibacterium and Brevibacterium) and the genus Micrococcus. Gram-negative bacteria, with the exception of some Acinetobacter spp., are generally not isolated from the skin, but are thought to arise in cultures owing to contamination from the gastrointestinal tract4.
Non-bacterial microorganisms have also been isolated from the skin. The most commonly isolated fungal species are Malassezia spp., which are especially prevalent in sebaceous areas. The Demodex mites (such as Demodex folliculorum and Demodex brevis), which are microscopic arthropods, are also regarded as part of the normal skin flora. Demodex mites feed on sebum and are more prevalent following puberty, preferring to colonize sebaceous areas of the face3. Demodex mites may also feed on epithelial cells lining the pilosebaceous unit, or even on other organisms (such as Propionibacterium acnes) that inhabit the same space. The role of commensal viruses has not been studied, and investigations are limited by the available molecular and microbiological means to identify and characterize viruses.
Historically, culture-based approaches have been the standard for characterizing microbial diversity. It is now evident that only a minority of bacteria are able to thrive in isolation103. Culture-based techniques essentially select for laboratory ‘weeds’: species that flourish under the typical nutritional and physiological conditions that are used by diagnostic microbiology laboratories. These are not necessarily the most abundant or influential organisms in the community. This bias is especially apparent when attempting to isolate skin microorganisms, which are inherently adapted to a cool, dry and acidic environment. Furthermore, hair follicles and sebaceous glands are anoxic environments and harbour anaerobic microorganisms. Isolation of anaerobes is particularly problematic using routine culture-based approaches104,105. These organisms are often slow growing and require special conditions for growth and during sample transport and processing.
The development of molecular techniques to identify and quantify microbial organisms has revolutionized our view of the microbial world. Genomic characterization of bacterial diversity relies on sequence analysis of the 16S ribosomal RNA gene, which is present in all bacteria and archaea but not in eukaryotes. The 16S rRNA gene contains species-specific hypervariable regions, which allow taxonomic classification, and highly conserved regions, which act as a molecular clock and a binding site for PCR primers106. The advent of new sequencing technologies (such as pyrosequencing) has massively increased throughput while decreasing the cost of sequencing. Importantly, an organism does not need to be cultured to determine its type by 16S rRNA sequencing.
Variation by skin site
Molecular approaches examining bacterial diversity have underlined the concept that the skin microbiota is dependent on the body site and that caution should be taken when selecting and comparing sites for skin microbiome studies. Our group and others have demonstrated that colonization of bacteria is dependent on the physiology of the skin site, with specific bacteria being associated with moist, dry and sebaceous microenvironments (FIG. 3). In general, bacterial diversity seems to be lowest in sebaceous sites, suggesting that there is selection for specific subsets of organisms that can tolerate conditions in these areas. Sebaceous sites that contain low phylotype richness include the forehead (six phylotypes43), the retroauricular crease (behind the ear) (15 phylotypes42), the back (17 phylotypes42) and the alar crease (side of the nostril) (18 phylotypes42). Propionibacterium spp. are the dominant organisms in these and other sebaceous areas, which confirms classical microbiological studies that describe Propionibacterium spp. as lipophilic residents of the pilosebaceous unit. Microbial transplant experiments suggest that the microenvironment of sebaceous areas (such as the forehead) is a stronger force in determining microbial colonization than the microenvironment of dry areas (such as the forearm)43.
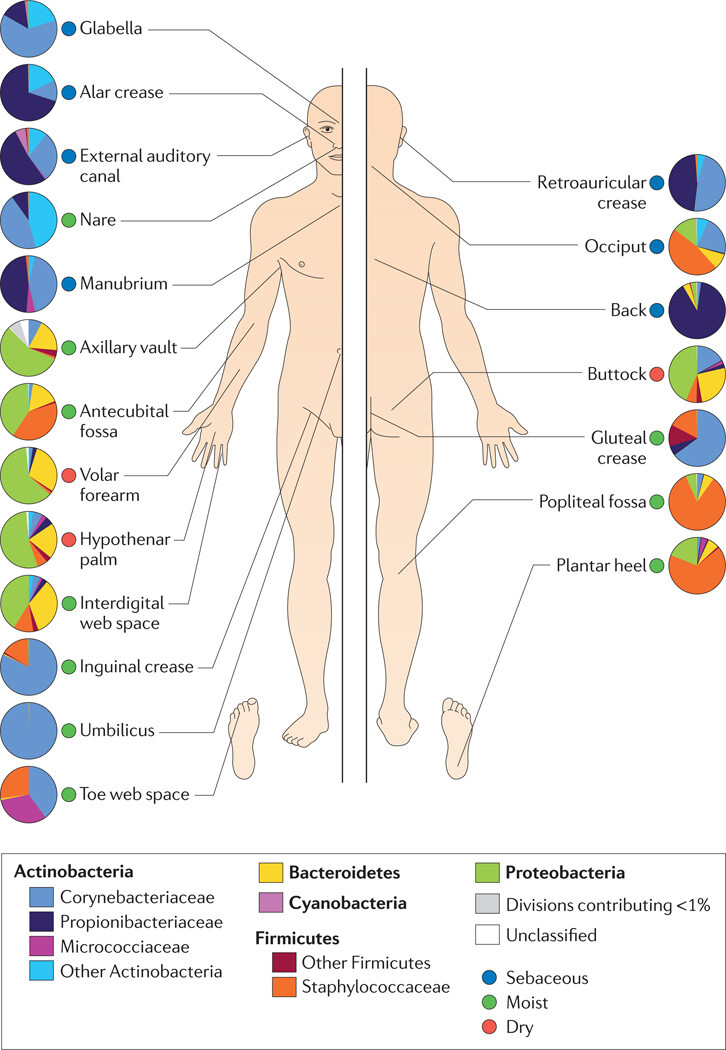
Figure 3
Topographical distribution of bacteria on skin sites
The skin microbiome is highly dependent on the microenvironment of the sampled site. The family-level classification of bacteria colonizing an individual subject is shown, with the phyla in bold. The sites selected were those that show a predilection for skin bacterial infections and are grouped as sebaceous or oily (blue circles), moist (typically skin creases) (green circles) and dry, flat surfaces (red circles). The sebaceous sites are: glabella (between the eyebrows); alar crease (side of the nostril); external auditory canal (inside the ear); retroauricular crease (behind the ear); occiput (back of the scalp); manubrium (upper chest); and back. Moist sites are: nare (inside the nostril); axillary vault (armpit); antecubital fossa (inner elbow); interdigital web space (between the middle and ring fingers); inguinal crease (side of the groin); gluteal crease (topmost part of the fold between the buttocks); popliteal fossa (behind the knee); plantar heel (bottom of the heel of the foot); toe web space; and umbilicus (navel). Dry sites are: volar forearm (inside of the mid-forearm); hypothenar palm (palm of the hand proximal to the little finger); and buttock. Data from 42.
Metagenomic analysis has revealed that Staphylococcus and Corynebacterium spp. are the most abundant organisms colonizing moist areas42,43, consistent with culture data suggesting that these organisms prefer areas of high humidity. These moist sites include the umbilicus (navel), the axillary vault, the inguinal crease (side of the groin), the gluteal crease (topmost part of the fold between the buttocks), the sole of the foot, the popliteal fossa (behind the knee) and the antecubital fossa (inner elbow). Staphylococci occupy an aerobic niche on the skin and probably use the urea present in sweat as a nitrogen source. Corynebacteria are extremely fastidious and slow-growing organisms in culture, and, as such, their role as skin microorganisms has been underappreciated until recently. Processing of apocrine sweat by corynebacteria and staphylococci (along with other axillary vault microorganisms) results in the characteristic malodour associated with sweat in humans15,16,18,50.
The most diverse skin sites are the dry areas, with mixed representation from the phyla Actinobacteria, Proteobacteria, Firmicutes and Bacteriodetes40,42,43. These sites include the forearm, buttock and various parts of the hand. A surprising feature of the microbiota of these sites is the abundance of Gram-negative organisms that are captured by molecular analysis; these were once thought to colonize the skin only rarely, as contaminants from the gastrointestinal tract1,4. Interestingly these sites also harbour greater phylogenetic diversity than the gut or the oral cavity of the same individual43.
Temporal variation
Molecular analysis of skin microbiota has also revealed that the temporal variability of the skin microbiome is dependent on the site sampled. The most consistent sites over time, when considering bacterial community membership and community structure, were the external auditory canal (inside the ear), the nare (inside the nostril) and the inguinal crease42. These are all sites that are at least partially occluded. In general, sites that harbour a greater diversity of microorganisms tend to be less stable over time in terms of community membership and structure; these sites include the volar forearm (forearm), the popliteal fossa, the antecubital fossa, the plantar heel (the bottom of the heel of the foot) and the interdigital web space (between the fingers) 42. Compared with the microbiome of the gut and the mouth, the microbiome of the skin had the greatest variability over time43
Interpersonal variation
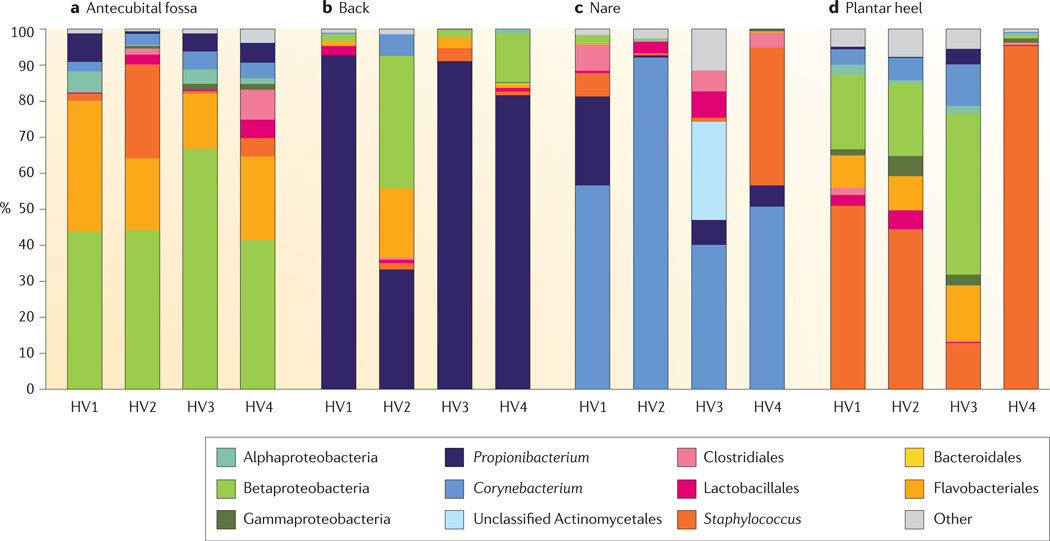
In general, intrapersonal variation in microbial community membership and structure between symmetric skin sites is less than the interpersonal variation, as determined by 16S rRNA metagenomic sequencing40,42,43. The bacteria that populate the sebaceous back are predominantly Propionibacterium spp., with some representation from the Betaproteobacteria (phylum Proteobacteria) and the Flavobacteriales (phylum Bacteroidetes). Although all three of these bacterial taxa are also found on the antecubital fossa and the plantar heel, the relative amounts of Propionibacterium spp. are much lower at these sites, which have a greater abundance of Betaproteobacteria and Staphylococcus spp., respectively. As shown in FIG. 4, the antecubital fossa, back, nare and plantar heel are more similar to the same site on another individual than to any other site on the same individual. In this sense, the ecological body site niche is a greater determinant of the microbiota composition than the individual genetic variation among healthy volunteers (FIG. 4). Some sites on an individual are similar, but these are sites such as the antecubital fossa and popliteal fossa that share common ecological features.
Figure 4
Interpersonal variation of the skin microbiome
The microbial distribution of four sites on four healthy volunteers (HV1, HV2, HV3 and HV4) is depicted at the antecubital fold (inner elbow; part a); the back (part b); the nare (inside the nostril; part c); and the plantar heel (bottom of the heel of the foot; part d). Skin microbial variation is more dependent on the site than on the individual. Bars represent the relative abundance of bacterial taxa as determined by 16S ribosomal RNA sequencing. Data from REF. 42.
A study with larger numbers of patients will be required to statistically define which bacterial species are unique to certain individuals or body sites. In a study of six subjects, it was shown that only 6.6% of all genera identified were found on the forearms of all subjects sampled, and 68.1% of genera identified were present on only one subject40. In a study of hand microbiota, hands from the same individual shared only 17% of species-level phylotypes, whereas hands from different individuals shared only 13%33. The frequency of hand washing, the degree of exposure to environment elements and handedness preference probably all contribute to the variation that is seen in the skin microbiota of the hand. Comparisons of male and female skin microbiota suggest that females harbour a greater diversity of bacteria on their hands, but it remains unclear whether this observed difference is due to physiological factors or differences in hygiene and cosmetic usage33,40. Our work has demonstrated that interpersonal variation, as measured by shared community membership and structure, is lowest in sebaceous areas such as the alar crease, the back and the manubrium (upper chest)42. These studies depict the skin microbiota as predominately composed of a handful of stable inhabitants (Propionibacterium and Staphylococcus spp.). Rare and/or transient species make up the balance and account for interpersonal variation. Factors driving the variability are unclear, but we suggest that they may include external environmental factors (for example, climate and geography), host immune status, host pathophysiology and/or historical exposures.
Further challenges: defining the functional potential of the skin microbiome
Although molecular approaches to surveying bacterial diversity provide a less biased depiction of skin microbiota than culture-based assays, they are not without their caveats. The molecular approaches that are currently in use are unable to distinguish between 16S rRNA genes that are derived from living versus dead organisms. Therefore, molecular studies are technically surveying the history of the skin microbiota. As the skin is an organ that is exposed to the environment, it is difficult to determine which of the identified species are transient and which species are resident members of the community. In addition, biases exist in the methods that are used to extract, amplify and sequence 16S rRNA genes. Even degenerate PCR primers may not amplify all 16S rRNA genes with equal efficiency. Furthermore, typing of 16S rRNA genes does not provide information regarding the gene content of flexible, open pan-genomes or plasmids. For example, 16S rRNA gene phylotyping cannot distinguish between methicillin-sensitive and methicillin-resistant Staphylococcus spp. isolates.
Only whole-genome shotgun metagenomic sequencing (WGS metagenomic sequencing) of bacterial communities will define their full genetic diversity and enable prediction of the gene functions that are associated with the skin microbiota. WGS metagenomic sequencing has proved to be useful in identifying the functional potential of gut microbiomes, revealing the increased energy harvesting potential of the obesity-associated gut microbiota51. To date, WGS metagenomic analysis of skin microbiota has not been reported. Several factors delay this type of analysis of skin microbiota. One such factor is the lack of reference genome sequences for skin isolates; this is likely to be due to the difficulty in culturing some skin microorganisms such as the corynebacteria. Furthermore, obtaining the critical amount of starting material required for WGS metagenomic sequencing, free of contaminating host DNA, is challenging for skin. Before skin metagenomic sequencing is tractable on a large scale, robust methods need to be further developed to separate host DNA from microorganism DNA, followed by unbiased whole-genome amplification.
Beyond the bacterial microbiome
Molecular approaches have been used to characterize eukaryotic species colonizing the skin, although these methods are not as well developed. The fungal phylogeny has been established using data from six genes: 18S rRNA, 28S rRNA, 5.8S rRNA, elongation factor 1α and two RNA polymerase II subunits52. When performing molecular surveys of fungal diversity, the most commonly analysed region is the internal transcribed spacer region that separates the small- and large-subunit rRNA genes in eukaryotes53. Most fungal organisms identified on the healthy skin by molecular typing resemble Malassezia spp., closely mirroring culture-based data54–56. In one study, Malassezia spp. were calculated to constitute 53–80% of the total skin fungal population, depending on the skin site, with the retroauricular crease harbouring the highest proportion56. It still remains unclear which fungal species constitute the remainder of the population, and further investigation is warranted. Culture-based analysis suggests that Candida spp. rarely colonize human skin but can cause clinical infection especially in conditions of immune deficiency, diabetes or infection following antibiotic use4,57. Other types of fungi that, according to culture-based analyses, are thought to grow on the skin, include Debaryomyces and Cryptococcus spp.3, although this has not been confirmed by molecular analysis of skin fungal biota. Larger, more extensive studies of the human skin fungi are needed, as well as better approaches for identifying and analysing fungal sequences.
Demodex mites (such as Demodex folliculorum and Demodex brevis), which are small arthropods, have historically been associated with rosacea, as well as a range of other skin disorders, including facial itching and chronic blephartitis58–60. Demodex mites reside in the pilosebaceous units, most commonly of the facial skin, and are considered part of the normal skin microflora60,61. Molecular methods for typing Demodex mites do not exist. Methods for isolating and identifying viruses from skin are just being developed, such as rolling-circle amplification62. These microorganisms are likely to be an important component of the skin ecosystem; thus, further analysis of their diversity and colonization dynamics are warranted (FIG. 1). To our knowledge, archaea have not been identified on the skin, either by culture or by molecular analyses.
Modulation by the cutaneous immune system
In addition to being a physical barrier, the skin is an immunological barrier63. The skin immune response is vital in wounding and infection and also modulates the commensal microbiota that colonizes the skin. Keratinocytes continuously sample the microbiota colonizing the skin surface through pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs), mannose receptors and the NOD-like receptors. These receptors recognize pathogen-associated molecular patterns (PAMPs) including flagellin and nucleic acids, as well as lipopolysaccharide from Gram-negative bacteria, mannan and zymosin from fungal cell walls, and peptidoglycan and lipoteichoic acid from Gram-positive bacteria. The activation of keratinocyte PRRs by PAMPs immediately initiates the innate immune response, resulting in the secretion of antimicrobial peptides (AMPs), cytokines and chemokines. Beyond effecting an adaptive immune response, AMPs also directly kill bacteria, fungi and enveloped viruses64. Therefore, there is a constant interplay among keratinocytes, immune cells and microorganisms that is modulated by AMPs, cytokines, chemokines and microbial peptides.
Despite being constantly exposed to large numbers of microorganisms, the skin can discriminate between harmless commensal microorganisms and harmful pathogenic microorganisms. The mechanism of this discrimination is not fully clear, but may involve the induction of immune tolerance; TLRs may be desensitized by prolonged exposure to commensal microorganisms, either through decreased TLR expression on the cell surface or by activation of the TLR pathway inhibitors interleukin-1 receptor-associated kinase 3 (IRAK3; also known as IRAK-M) and suppressor of cytokine signalling 1 (SOCS1)65,66. Specificity may also be achieved by combined recognition of PAMPs by PRRs.
Staphylococcus epidermidis, a commensal bacterium, has recently been demonstrated to modulate the host innate immune response. Phenol-soluble modulins produced by S. epidermidis can selectively inhibit skin pathogens, such as S. aureus and Group A Streptococcus, and can even co-operate with host AMPs to enhance killing67,68. Recent studies demonstrate that commensal-induced TLR signalling may be necessary for cell survival and repair during infection. Lipoteichoic acid produced by S. epidermidis can inhibit skin inflammation through a TLR2- and TLR3-mediated crosstalk mechanism69. In addition, S. epidermidis triggers keratinocyte expression of AMPs through a TLR2-dependent mechanism70. This body of work definitively links commensal skin microorganisms with modulation of the innate immune response.
Dysregulation of the skin immune response is apparent in several skin disorders (for example, psoriasis, atopic dermatitis (AD; commonly known as eczema and contact dermatitis), but how dysregulation affects and/or results from changes in the microbiota remains unclear. AD lesions are characterized by low levels of AMP production as compared with levels from normal skin. This is in sharp contrast to psoriatic lesions, which produce abundant quantities of AMPs and are characterized by an activated innate immune response71–75. AD lesions are also regularly infected with pathogens, especially S. aureus, and respond to antimicrobial treatment. Upregulation of T helper 2 cytokines in AD lesional skin is likely to partially account for the apparent suppression of the innate immune response that is observed in AD71,72. There is no clear microbial component to the common form of psoriasis, although the guttate subset of psoriasis has been associated with streptococcal infections76.
The skin microbiome and disease
Many common skin disorders are postulated to have an underlying microbial contribution because clinical improvement is seen with antimicrobial treatments. However, a causative microbial component that fully satisfies Koch’s postulates has rarely been identified in these skin diseases. We illustrate the different ways in which a skin disease can be associated with a specific organism with three cases: first, skin disorders with a correlation to microbiota; second, a skin disorder with a currently unidentified microbial component; and third, a skin commensal that can become invasive to cause infection.
Skin disorders with a correlation to microbiota
Seborrhoeic dermatitis is a hyperproliferative, pruritic skin disorder, typically affecting the scalp. A fungal component is postulated to participate in disease pathogenesis, as a wide range of fungicides effectively combat seborrhoeic dermatitis.The presumptive target of these fungicides is Malassezia spp., which are the dominant fungi cultured from the skin and are particularly prevalent in sebaceous areas such as the scalp. Improvements in seborrhoeic dermatitis are associated with reduced levels of Malassezia spp. on the scalp77. However, improvement is not observed when the scalp is treated with antibacterial agents78. The suggested mechanism for improvement implicates Malassezia lipase genes, which process sebum to release free fatty acid metabolites (oleic acid). These metabolites then penetrate upper layers of the skin to promote hyperproliferation and inflammation79. As Malassezia spp. are present on healthy skin, and by themselves are not sufficient to cause seborrhoeic dermatitis, other factors probably contribute to their pathogenicity and ability to cause disease.
The commensal skin bacterium P. acnes is associated with the very common teenage malady acne, an inflammatory disorder of the pilosebaceous unit. The onset of puberty matures the pilosebaceous unit, increasing the preponderance of lipophilic microorganisms, especially P. acnes, which secretes lipases, proteases and hyaluronidases that injure the tissue lining of the pilosebaceous unit80. The P. acnes genome encodes various immunogenic factors, including cell surface proteins with adherent properties and porphyrins19. Furthermore, the damage caused by P. acnes in the pilosebaceous unit activates the classical and alternative complement pathways81,82 and induces production of pro-inflammatory cytokines83,84 and neutrophil chemotactic factors85,86. However, the use of molecular methods to examine the bacterial component of acne has been limited, and a small sequencing study did not identify any novel acne-associated bacteria87.
AD is a chronic relapsing disorder that affects ~15% of US children and ~2% of adults, and is also associated with microbial colonization and infection. The prevalence of AD has doubled or tripled in industrialized countries over the past three decades with no clear cause. This raises the intriguing possibility that skin microbial fluctuations modulate the gene–environment interaction on the skin surface, resulting in the episodic exacerbations of AD. Classic AD manifests at stereotypical sites, including the antecubital fossa and the popliteal fossa, which are sites that harbour similar organisms when compared to other body sites42. More than 90% of AD patients are colonized with S. aureus on both lesional and non-lesional skin, compared with <5% of healthy individuals88,89. The most common treatments for AD include topical or systemic antibiotics, and steroids. Dilute bleach baths to lower the bacterial load are effective in reducing clinical severity90. No specific link has been identified between virulence factors expressed by S. aureus and flare-ups in patients with AD. However, in a mouse model that has reduced skin barrier function (the NC/Nga strain, which is deficient in ceramide production91), application of S. aureus immunoglobulin G-binding protein A (also known as Staphylococcus protein A) along with an agitating detergent resulted in a severe AD-like phenotype92.
Disorder with an unidentified microbial component
Chronic wounds, affecting diabetic, elderly, and immobile individuals, are an example where commensal skin organisms invade and become pathogenic upon breach of the skin barrier. Although bacteria do not cause the initial wounding event, they are thought to contribute to the lack of healing and persistent inflammation that is associated with chronic wounds. As for healthy skin, the microbial diversity colonizing chronic wounds is greater when analysed using molecular methods as opposed to culture-based methods93. However, molecular studies thus far have been unable to identify a unique organism that colonizes wounds of the same aetiology (for example, in diabetic foot ulcers or venous leg ulcers)94–96. This is in contrast to burn wounds, in which a causative microbiological agent is usually readily identifiable. Burn wounds commonly become infected with S. pyogenes, Enterococcus spp. or Pseudomonas aeruginosa, and can also become infected with fungi and/or viruses97.
A major problem in dissecting the clinical relevance of chronic wound microbial diversity data sets is the environmental and genetic factors that may confound results, especially when the precise clinical phenotyping data that could be used to stratify patient populations are absent. One way to circumvent confounding factors when analysing host–microorganism relationships is to use an animal model. This approach has allowed the identification of a longitudinal selective shift in the microbiota colonizing slow-healing diabetic mouse wounds; the shift is correlated with aberrant expression of skin defence and inflammatory genes98.
S. epidermidis: an invasive skin commensal that causes infection
The third category is that of skin microorganisms that are normally commensal but that can sometimes cause infection and disease, especially when they invade other sites. S. epidermidis is a very common skin commensal, but it is also the most frequent cause of hospital- acquired infection on in-dwelling medical devices such as catheters or heart valves99. After they gain entry, virulent strains of these organisms can form biofilms on catheters or other devices, which protects them from the host immune system and antibiotics. Increasing levels antibiotic resistance, particularly to oxacillin or methicillin, complicates treatment of S. epidermidis infections. Furthermore, S. epidermidis seems to be a reservoir of antibiotic-resistance genes that it transfers to the closely related but more virulent organism, S. aureus100.
Conclusions and perspectives
Molecular approaches to characterizing microbial diversity have dramatically changed our view of the skin microbiome, subsequently raising many important questions about the host–microorganism relationship and its relevance to skin disease. Although it is now clear that several dominant organisms (that is, Staphylococcus and Propionibacterium spp.) constitute a large proportion of the skin microbiota, little is understood about the rare or transient organisms making up the balance. It is unclear what factors drive variation in these organisms, and how fluctuation is associated with skin disease. Metagenomic analysis to elucidate the full complement of microbial genes and their functions should provide insight into these questions. The US National Institutes of Health Common Fund Human Microbiome Project aims to characterize the human microbiota and its role in health by examining the microbial diversity of 250 healthy volunteers sampled at 18 skin sites, including two skin sites, nine oral cavity sites, the nare, the stool and five vaginal samples for women101. These results from healthy volunteers will be used to guide and examine the statistical power of clinical studies, including investigations into the skin disorders acne, AD and psoriasis.
Although there is a general agreement that microorganisms are potential components of many skin disorders, including those described here, little is understood about their contribution and how it relates to the genetic and environmental variation that also contributes to the disease. Many common skin diseases are associated with a specific stage of life, a specific topographical location and/or specific microorganisms. Whether this specificity is driven by the endogenous microbial community structure remains to be determined. Several skin sites with a predilection for disease, such as the glans penis and the eyelids, remain to be sampled and characterized for bacterial diversity.
Another outstanding question is whether indigenous skin microorganisms provide some benefit to the host, and whether they are truly symbiotic, or commensal. In a recent example of host and microorganism joining forces to combat invasion by pathogens, the commensal skin bacteria S. epidermidis was demonstrated to inhibit nare colonization and biofilm formation by S. aureus102. A subset of S. epidermidis express the glutamyl endopeptidase protein (encoded by the esp gene), which can synergize with the human AMP β-defensin 2 (also known as β-defensin 4A) to interfere with S. aureus colonization. This example raises several important points for consideration, including the possibility of the host and the microorganism evolving together. Furthermore, as our arsenal of antimicrobial weapons falls short in the battle against S. aureus and other potential pathogens, perhaps therapeutics derived from microorganisms themselves will offer promise as viable alternatives. As we continue to enhance our knowledge of the skin microbiota, these factors and other unanswered questions will guide future research efforts directed towards understanding the complex interactions governing the host–microorganism relationship.
Acknowledgements
We thank H. Kong and E. Hobbs for critical reading of the manuscript and J. Fekecs and D. Leja for graphical assistance. E.A.G. is supported by a Pharmacology Research Associate Training Fellowship, US National Institute of General Medical Sciences. This work was supported by the US National Human Genome Research Institute Intramural Research Program and the US National Institutes of Health Common Fund AR057504.
Glossary
| Keratinocyte | The predominant cell type of the epidermis. Keratinocytes produce keratin as they terminally differentiate into the squames of the stratum corneum |
| Squame | An enucleated, dead, squamous keratinocyte that is shed from the stratum corneum |
| Sebum | The oily, lipid-containing substance that is secreted by the sebaceous glands of the skin. Sebaceous glands are connected to the hair follicle and form the pilosebaceous unit. Sebum protects and emolliates the skin and hair |
| 16S ribosomal RNA metagenomic sequencing | Genomic analysis of 16S ribosomal RNA phylotypes from DNA that is extracted directly from bacterial communities in clinical or environmental samples, a process that circumvents culturing |
| Microbiome | All of the genetic material of a microbial community sequenced together |
| Phylotype | A taxon-neutral way to describe organisms based on their phylogenetic relationships to other organisms. Phylotypes are determined by comparing 16S ribosomal RNA gene sequences. A common threshold used to define species-level phylotypes is 97% sequence identity of the 16S rRNA gene sequence |
| Whole-genome shotgun metagenomic sequencing | Genomic analysis of DNA that is extracted directly from a clinical or environmental sample and whole-genome shotgun (WGS) sequenced to represent the full microbiome |
| Pattern recognition receptor (PRR) | A receptor present on the surface of keratinocytes and other cells of the innate immune system that recognizes microorganism-specific molecules (for example, lipopolysaccharide and flagellin) |
| Pathogen-associated molecular pattern (PAMP) | A molecule that is associated with a pathogen and recognized by a pathogen recognition receptor. Examples include lipopolysaccharide, flagellin, lipoteichoic acid, double-stranded RNA, peptidoglycan and unmethylated CpG motifs |
| Atopic dermatitis (AD) | A type of eczema characterized by red, flaky, itchy skin, typically affecting the inner elbows and behind the knees. It is often associated with other atopic diseases such as allergic rhinitis, hay fever and asthma |
| Seborrhoeic dermatitis | An inflammatory, hyperproliferative skin condition characterized by red, flaky, skin often affecting sebaceous areas of the face, scalp and trunk. Commonly known as dandruff |
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- Chiller K, Selkin BA, Murakawa GJ. Skin microflora and bacterial infections of the skin. J. Investig. Dermatol. Symp. Proc. 2001;6:170–174.
- Fredricks DN. Microbial ecology of human skin in health and disease. J. Investig. Dermatol. Symp. Proc.
- The Ecology of the Human Skin. Bannerstone House, Springfield, Illinois: Charles C Thomas; 1965. A seminal and comprehensive work of classical dermatological microbiology.
- Roth RR, James WD. Microbial ecology of the skin. Annu. Rev. Microbiol. 1988;42:441–464.
- Noble WC. Skin microbiology: coming of age. J. Med. Microbiol. 1984;17:1–12.
- Roth RR, James WD. Microbiology of the skin: resident flora, ecology, infection. J. Am. Acad. Dermatol. 1989;20:367–390.
- Cogen AL, Nizet V, Gallo RL. Skin microbiota: a source of disease or defence? Br. J. Dermatol. 2008;158:442–455.
- Tagami H. Location-related differences in structure and function of the stratum corneum with special emphasis on those of the facial skin. Int. J. Cosmet Sci. 2008;30:413–434.
- Proksch E, Brandner JM, Jensen JM. The skin: an indispensable barrier. Exp. Dermatol. 2008;17:1063–1072.
- Elias PM. The skin barrier as an innate immune element. Semin. Immunopathol. 2007;29:3–14.
- Segre JA. Epidermal barrier formation and recovery in skin disorders. J. Clin. Invest. 2006;116:1150–1158.
- Fuchs E, Raghavan S. Getting under the skin of epidermal morphogenesis. Nature Rev. Genet. 2002;3:199–209.
- Leeming JP, Holland KT, Cunliffe WJ. The microbial ecology of pilosebaceous units isolated from human skin. J. Gen. Microbiol. 1984;130:803–807.
- Cohn BA. In search of human skin pheromones. Arch. Dermatol. 1994;130:1048–1051.
- Emter R, Natsch A. The sequential action of a dipeptidase and a β-lyase is required for the release of the human body odorant 3-methyl-3-sulfanylhexan-1-ol from a secreted Cys-Gly-( S ) conjugate by Corynebacteria. J. Biol. Chem. 2008;283:20645–20652.
- Decreau RA, Marson CM, Smith KE, Behan JM. Production of malodorous steroids from androsta-5,16-dienes and androsta-4,16-dienes by Corynebacteria and other human axillary bacteria. J. Steroid Biochem. Mol. Biol. 2003;87:327–336.
- Martin A, et al. A functional ABCC11 allele is essential in the biochemical formation of human axillary odor. J. Invest. Dermatol. 2010;130:529–540.
- Natsch A, Gfeller H, Gygax P, Schmid J, Acuna G. A specific bacterial aminoacylase cleaves odorant precursors secreted in the human axilla. J. Biol. Chem. 2003;278:5718–5727.
- Bruggemann H, et al. The complete genome sequence of Propionibacterium acnes a commensal of human skin. Science. 2004;305:671–673.
- Marples RR, Downing DT, Kligman AM. Control of free fatty acids in human surface lipids by Corynebacterium acnes. J. Invest. Dermatol. 1971;56:127–131.
- Ingham E, Holland KT, Gowland G, Cunliffe WJ. Partial purification and characterization of lipase (EC 3.1.1.3) from Propionibacterium acnes. J. Gen. Microbiol. 1981;124:393–401.
- Gribbon EM, Cunliffe WJ, Holland KT. Interaction of Propionibacterium acnes with skin lipids in vitro. J. Gen. Microbiol. 1993;139:1745–1751.
- Korting HC, Hubner K, Greiner K, Hamm G, Braun-Falco O. Differences in the skin surface pH and bacterial microflora due to the long-term application of synthetic detergent preparations of pH 5.5 and pH 7.0. Results of a crossover trial in healthy volunteers. Acta Derm. Venereol. 1990;70:429–431.
- Aly R, Shirley C, Cunico B, Maibach HI. Effect of prolonged occlusion on the microbial flora, pH, carbon dioxide and transepidermal water loss on human skin. J. Invest. Dermatol. 1978;71:378–381.
- Hentges DJ. The anaerobic microflora of the human body. Clin. Infect. Dis. 1993;16:S175–S180.
- Webster GF, Ruggieri MR, McGinley KJ. Correlation of Propionibacterium acnes populations with the presence of triglycerides on nonhuman skin. Appl. Environ. Microbiol. 1981;41:1269–1270.
- Leyden JJ, McGinley KJ, Mills OH, Kligman AM. Age-related changes in the resident bacterial flora of the human face. J. Invest. Dermatol. 1975;65:379–381.
- Somerville DA. The normal flora of the skin in different age groups. Br. J. Dermatol. 1969;81:248–258.
- Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA. 2010;107:11971–11975.
- Sarkany I, Gaylarde CC. Bacterial colonisation of the skin of the newborn. J. Pathol. Bacteriol. 1968;95:115–122.
- Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177.
- Marples RR. Sex, constancy, and skin bacteria. Arch. Dermatol. Res. 1982;272:317–320.
- Fierer N, Hamady M, Lauber CL, Knight R. The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc. Natl Acad. Sci. USA. 2008;105:17994–17999.
- Giacomoni PU, Mammone T, Teri M. Genderlinked differences in human skin. J. Dermatol. Sci. 2009;55:144–149.
- Dethlefsen L, Relman DA. Microbes and Health Sackler Colloquium: Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl Acad. Sci. USA. 2010 Sep 16;
- Antonopoulos DA, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 2009;77:2367–2375.
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280.
- McBride ME, Duncan WC, Knox JM. The environment and the microbial ecology of human skin. Appl. Environ. Microbiol. 1977;33:603–608.
- Faergemann J, Larko O. The effect of UV-light on human skin microorganisms. Acta Derm. Venereol. 1987;67:69–72.
- Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl Acad. Sci. USA. 2007;104:2927–2932.
- Grice EA, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324:1190–1192. A comprehensive analysis of skin microbiota across 20 sites.
- Costello EK, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. A comprehensive analysis of skin, gut and oral microbiota in the same individuals.
- Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638.
- Dewhirst FE, et al. The human oral microbiome. J. Bacteriol. 2010;192:5002–5017.
- Zaura E, Keijser BJ, Huse SM, Crielaard W. Defining the healthy ‘core microbiome’ of oral microbial communities. BMC Microbiol. 2009;9:259.
- Bik EM, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974.
- Pei Z, et al. Bacterial biota in the human distal esophagus. Proc. Natl Acad. Sci. USA. 2004;101:4250–4255.
- Bik EM, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl Acad. Sci. USA. 2006;103:732–737.
- Leyden JJ, McGinley KJ, Holzle E, Labows JN, Kligman AM. The microbiology of the human axilla and its relationship to axillary odor. J. Invest. Dermatol. 1981;77:413–416.
- Turnbaugh PJ, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. An important study demonstrating the functional potential of the human microbiome.
- James TY, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822.
- Chase MW, Fay MF. Ecology. Barcoding of plants and fungi. Science. 2009;325:682–683.
- Paulino LC, Tseng CH, Blaser MJ. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 2008;8:460–471.
- Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J. Clin. Microbiol. 2006;44:2933–2941.
- Gao Z, Perez-Perez GI, Chen Y, Blaser MJ. Quantitation of major human cutaneous bacterial and fungal populations. J. Clin. Microbiol. 2010;48:3575–3581.
- Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nature Rev. Microbiol. 2010;8:340–349. This review describes the clinical and molecular characteristics of bacterium–fungus interactions that are relevant to human disease with a focus on Candida spp.
- Lacey N, Delaney S, Kavanagh K, Powell FC. Mite-related bacterial antigens stimulate inflammatory cells in rosacea. Br. J. Dermatol. 2007;157:474–481.
- Georgala S, et al. Increased density of Demodex folliculorum and evidence of delayed hypersensitivity reaction in subjects with papulopustular rosacea. J. Eur. Acad. Dermatol. Venereol. 2001;15:441–444.
- Elston DM. Demodex mites: facts and controversies. Clin. Dermatol. 2010;28:502–504.
- Hay R. Demodex and skin infection: fact or fiction. Curr. Opin. Infect. Dis. 2010;23:103–105.
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe. 2010;7:509–515. An investigation of the preponderance of Merkel cell polyomavirus, and a methodology to isolate circular DNA viral genomes from human skin swabs.
- Borkowski AW, Gallo RL. The coordinated response of the physical and antimicrobial peptide barriers of the skin. J. Invest. Dermatol. 2011;131:285–287.
- Braff MH, Bardan A, Nizet V, Gallo RL. Cutaneous defence mechanisms by antimicrobial peptides. J. Invest. Dermatol. 2005;125:9–13.
- Strober W. Epithelial cells pay a Toll for protection. Nature Med. 2004;10:898–900.
- Fukao T, Koyasu S. PI3K and negative regulation of TLR signaling. Trends Immunol. 2003;24:358–363.
- Cogen AL, et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis a normal resident of the skin. J. Invest. Dermatol. 2010;130:192–200.
- Cogen AL, et al. Staphylococcus epidermidis antimicrobial δ -toxin (phenol-soluble modulin-γ) cooperates with host antimicrobial peptides to kill Group A Streptococcus. PLoS ONE. 2010;5:e8557.
- Lai Y, et al. Commensal bacteria regulate Toll-like receptor 3-dependent inflammation after skin injury. Nature Med. 2009;15:1377–1382. This analysis demonstrated that products of a skin commensal can modulate the innate immune response.
- Lai Y, et al. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defence against bacterial skin infections. J. Invest. Dermatol. 2010;130:2211–2221.
- Nomura I, et al. Distinct patterns of gene expression in the skin lesions of atopic dermatitis and psoriasis: a gene microarray analysis. J. Allergy Clin. Immunol. 2003;112:1195–1202.
- Nomura I, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J. Immunol. 2003;171:3262–3269.
- Gudjonsson JE, et al. Global gene expression analysis reveals evidence for decreased lipid biosynthesis and increased innate immunity in uninvolved psoriatic skin. J. Invest. Dermatol. 2009;129:2795–2804.
- Ong PY, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N. Engl. J. Med. 2002;347:1151–1160.
- de Jongh GJ, et al. High expression levels of keratinocyte antimicrobial proteins in psoriasis compared with atopic dermatitis. J. Invest. Dermatol. 2005;125:1163–1173.
- Owen CM, Chalmers RJ, O’Sullivan T, Griffiths CE. A systematic review of antistreptococcal interventions for guttate and chronic plaque psoriasis. Br. J. Dermatol. 2001;145:886–890.
- Pierard GE, Arrese JE, Pierard-Franchimont C, De Doncker P. Prolonged effects of antidandruff shampoos — time to recurrence of Malassezia ovalis colonization of skin. Int. J. Cosmet. Sci. 1997;19:111–117.
- Leyden JJ, McGinley KJ, Kligman AM. Role of microorganisms in dandruff. Arch. Dermatol. 1976;112:333–338.
- Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL., Jr Skin diseases associated with Malassezia species. J. Acad. Dermatol. 2004;51:785–798.
- Dessinioti C, Katsambas AD. The role of Propionibacterium acnes in acne pathogenesis: facts and controversies. Clin. Dermatol. 2010;28:2–7.
- Scott DG, Cunliffe WJ, Gowland G. Activation of complement — a mechanism for the inflammation in acne. Br. J. Dermatol. 1979;101:315–320.
- Webster GF, Leyden JJ, Nilsson UR. Complement activation in acne vulgaris: consumption of complement by comedones. Infect. Immun. 1979;26:183–186.
- Jeremy AH, Holland DB, Roberts SG, Thomson KF, Cunliffe WJ. Inflammatory events are involved in acne lesion initiation. J. Invest. Dermatol. 2003;121:20–27.
- Kim J. Review of the innate immune response in acne vulgaris: activation of Toll-like receptor 2 in acne triggers inflammatory cytokine responses. Dermatology. 2005;211:193–198.
- Puhvel SM, Sakamoto M. Cytotaxin production by comedonal bacteria (Propionibacterium acnes, Propionibacterium granulosum and Staphylococcus epidermidis ) J. Invest. Dermatol. 1980;74:36–39.
- Webster GF, Leyden JJ. Characterization of serum-independent polymorphonuclear leukocyte chemotactic factors produced by Propionibacterium acnes. Inflammation. 1980;4:261–269.
- Bek-Thomsen M, Lomholt HB, Kilian M. Acne is not associated with yet-uncultured bacteria. J. Clin. Microbiol. 2008;46:3355–3360.
- Hanifin JM, Rogge JL. Staphylococcal infections in patients with atopic dermatitis. Arch. Dermatol. 1977;113:1383–1386.
- Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. Br. J. Dermatol. 1974;90:525–530.
- Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–e814.
- Aioi A, et al. Impairment of skin barrier function in NC/Nga Tnd mice as a possible model for atopic dermatitis. Br. J. Dermatol. 2001;144:12–18.
- Terada M, et al. Contribution of IL-18 to atopic-dermatitis-like skin inflammation induced by Staphylococcus aureus product in mice. Proc. Natl Acad. Sci. USA. 2006;103:8816–8821.
- Frank DN, et al. Microbial diversity in chronic open wounds. Wound Repair Regen. 2009;17:163–172.
- Dowd SE, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008;8:43.
- Smith DM, et al. Evaluation of the bacterial diversity of Pressure ulcers using bTEFAP pyrosequencing. BMC Med. Genomics. 2010;3:41.
- Price LB, et al. Community analysis of chronic wound bacteria using 16S rRNA gene-based pyrosequencing: impact of diabetes and antibiotics on chronic wound microbiota. PLoS ONE. 2009;4:e6462.
- Polavarapu N, Ogilvie MP, Panthaki ZJ. Microbiology of burn wound infections. J. Craniofac. Surg. 2008;19:899–902.
- Grice EA, et al. Longitudinal shift in diabetic wound microbiota correlates with prolonged skin defence response. Proc. Natl Acad. Sci. USA. 2010;107:14799–14804. This study showed that a selective shift in microbiota is associated with an altered innate immune response.
- Uckay I, et al. Foreign body infections due to Staphylococcus epidermidis. Ann. Med. 2009;41:109–119.
- Otto M. Staphylococcus epidermidis — the ‘accidental’ pathogen. Nature Rev. Microbiol. 2009;7:555–567.
- Peterson J, et al. The NIH Human Microbiome Project. Genome Res. 2009;19:2317–2323. A detailed description of the Human Microbiome Project and its objectives.
- Iwase T, et al. Staphylococcus epidermidis esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. An important paper demonstrating the mechanism by which S. epidermidis inhibits S. aureus colonization of the nare.
- Dunbar J, Barns SM, Ticknor LO, Kuske CR. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 2002;68:3035–3045.
- Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin. Microbiol. Rev. 2001;14:244–269.
- Davies CE, et al. Use of molecular techniques to study microbial diversity in the skin: chronic wounds reevaluated. Wound Repair Regen. 2001;9:332–340.
- Hugenholtz P, Pace NR. Identifying microbial diversity in the natural environment: a molecular phylogenetic approach. Trends Biotechnol. 1996;14:190–197.
Nat Rev Microbiol. Author manuscript; available in PMC 2013 Jan 3.
Published in final edited form as:
Nat Rev Microbiol. 2011 Apr; 9(4): 244–253.
doi: 10.1038/nrmicro2537
PMCID: PMC3535073
NIHMSID: NIHMS424100
PMID: 21407241
Elizabeth A. Grice and Julia A. Segre